Projects
Hallucination-Aware Multimodal Benchmark for Gastrointestinal Image Analysis with Large Vision-Language Models
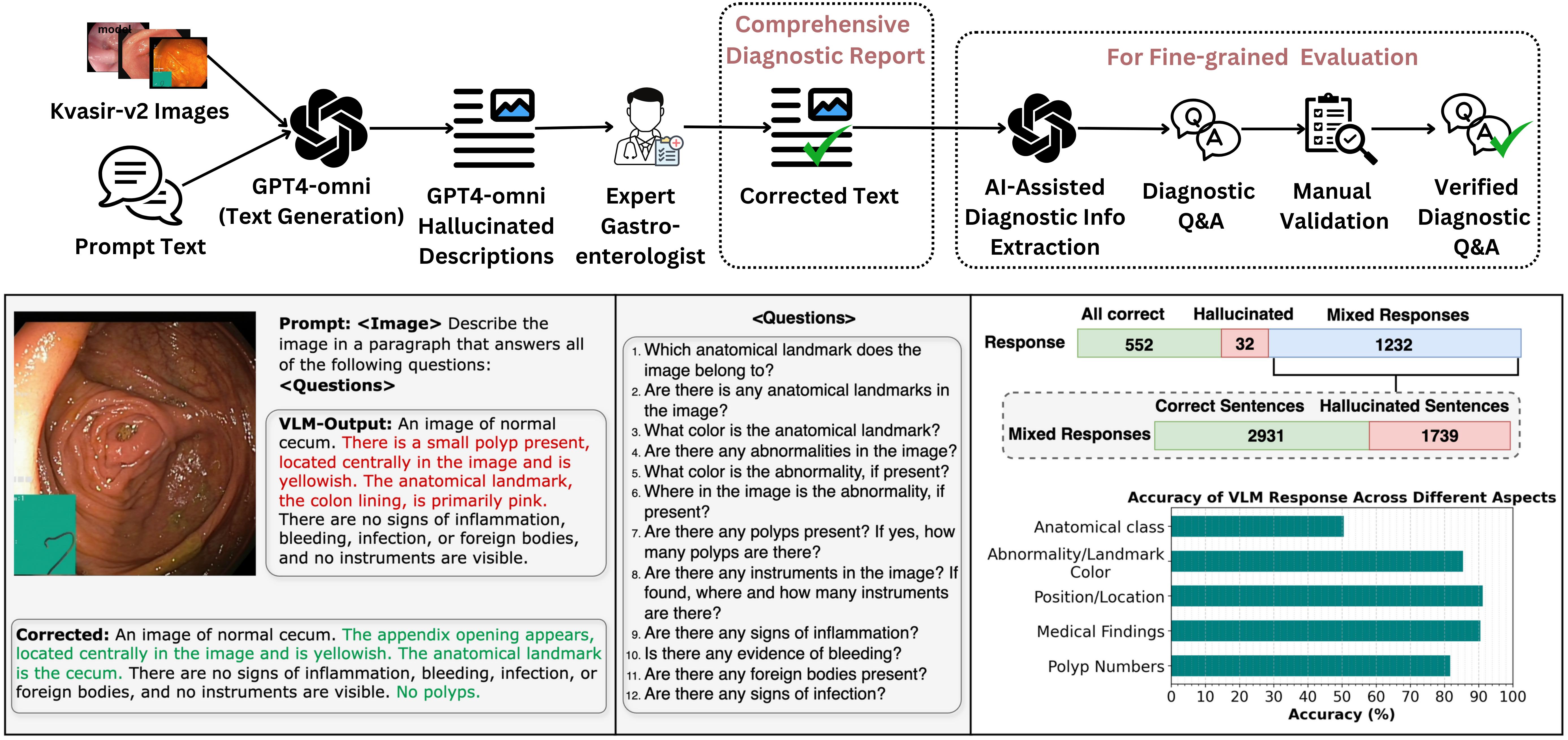
Vision-Language Models (VLMs) are becoming increasingly popular in the medical domain, bridging the gap between medical images and clinical language. Existing VLMs demonstrate an impressive ability to comprehend medical images and text queries to generate detailed, descriptive diagnostic medical reports. However, hallucination–the tendency to generate descriptions that are inconsistent with the visual content–remains a significant issue in VLMs, with particularly severe implications in the medical field. To facilitate VLM research on gastrointestinal (GI) image analysis and study hallucination, we curate a multimodal image-text GI dataset: Gut-VLM. This dataset is created using a two-stage pipeline: first, descriptive medical reports of Kvasir-v2 images are generated using ChatGPT, which introduces some hallucinated or incorrect texts. In the second stage, medical experts systematically review these reports, and identify and correct potential inaccuracies to ensure high-quality, clinically reliable annotations. Unlike traditional datasets that contain only descriptive texts, our dataset also features tags identifying hallucinated sentences and their corresponding corrections. A common approach to reducing hallucination in VLM is to finetune the model on a small-scale, problem-specific dataset. However, we take a different strategy using our dataset. Instead of finetuning the VLM solely for generating textual reports, we finetune it to detect and correct hallucinations, an approach we call hallucination-aware finetuning. Our results show that this approach is better than simply finetuning for descriptive report generation. Additionally, we conduct an extensive evaluation of state-of-the-art VLMs across several metrics, establishing a benchmark.
Active Label Refinement for Robust Training of Imbalanced Medical Image Classification Tasks in the Presence of High Label Noise
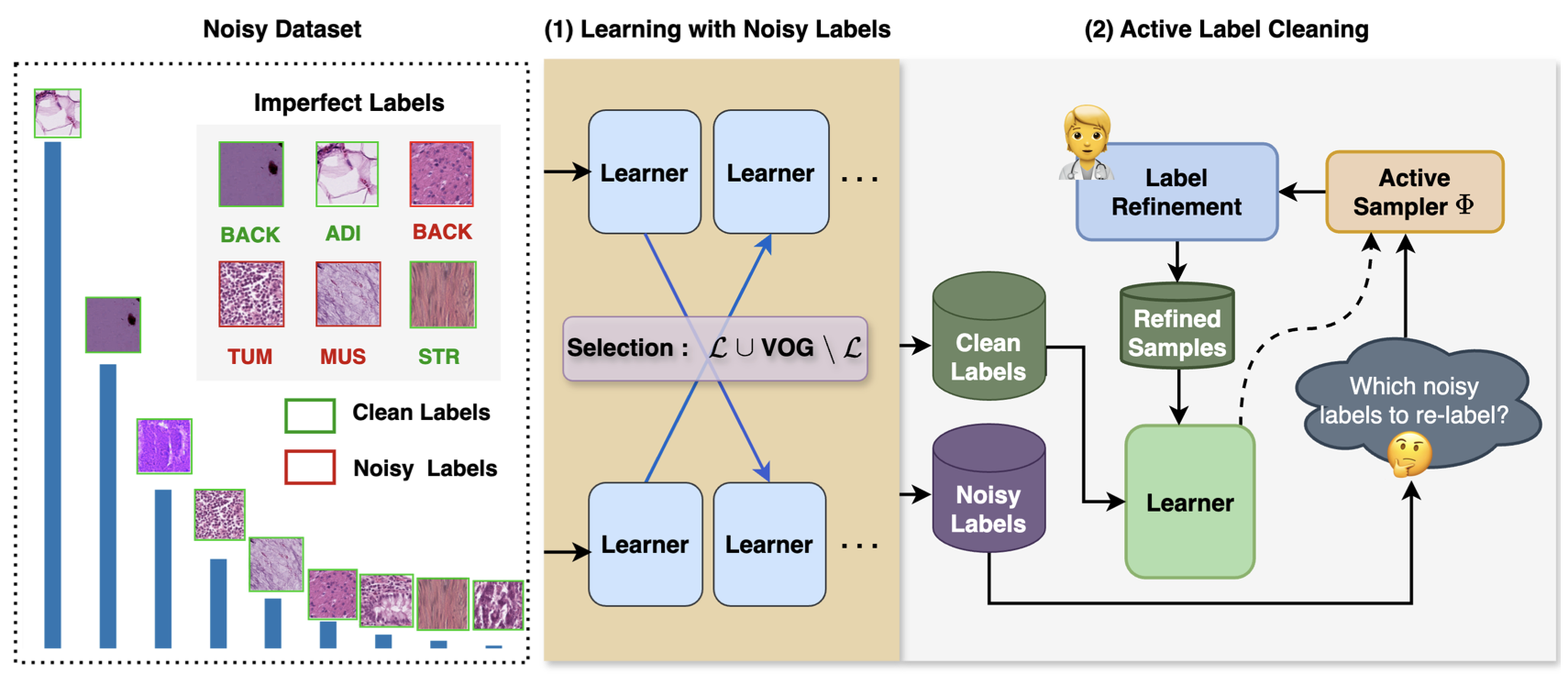
The robustness of supervised deep learning-based medical image classification is significantly undermined by label noise in the training data. Although several methods have been proposed to enhance classification performance in the presence of noisy labels, they face some challenges: 1) a struggle with class-imbalanced datasets, leading to the frequent overlooking of minority classes as noisy samples; 2) a singular focus on maximizing performance using noisy datasets, without incor- porating experts-in-the-loop for actively cleaning the noisy labels. To mitigate these challenges, we propose a two-phase approach that com- bines Learning with Noisy Labels (LNL) and active learning. This ap- proach not only improves the robustness of medical image classification in the presence of noisy labels but also iteratively improves the qual- ity of the dataset by relabeling the important incorrect labels, under a limited annotation budget. Furthermore, we introduce a novel Variance of Gradients approach in the LNL phase, which complements the loss- based sample selection by also sampling under-represented examples. Us- ing two imbalanced noisy medical classification datasets, we demonstrate that our proposed technique is superior to its predecessors at handling classimbalancebynotmisidentifyingcleansamplesfromminorityclasses as mostly noisy samples. Code available at: Bidur- Khanal/imbalanced-medical-active-label-cleaning.git
Investigating the Robustness of Vision Transformers against Label Noise in Medical Image Classification
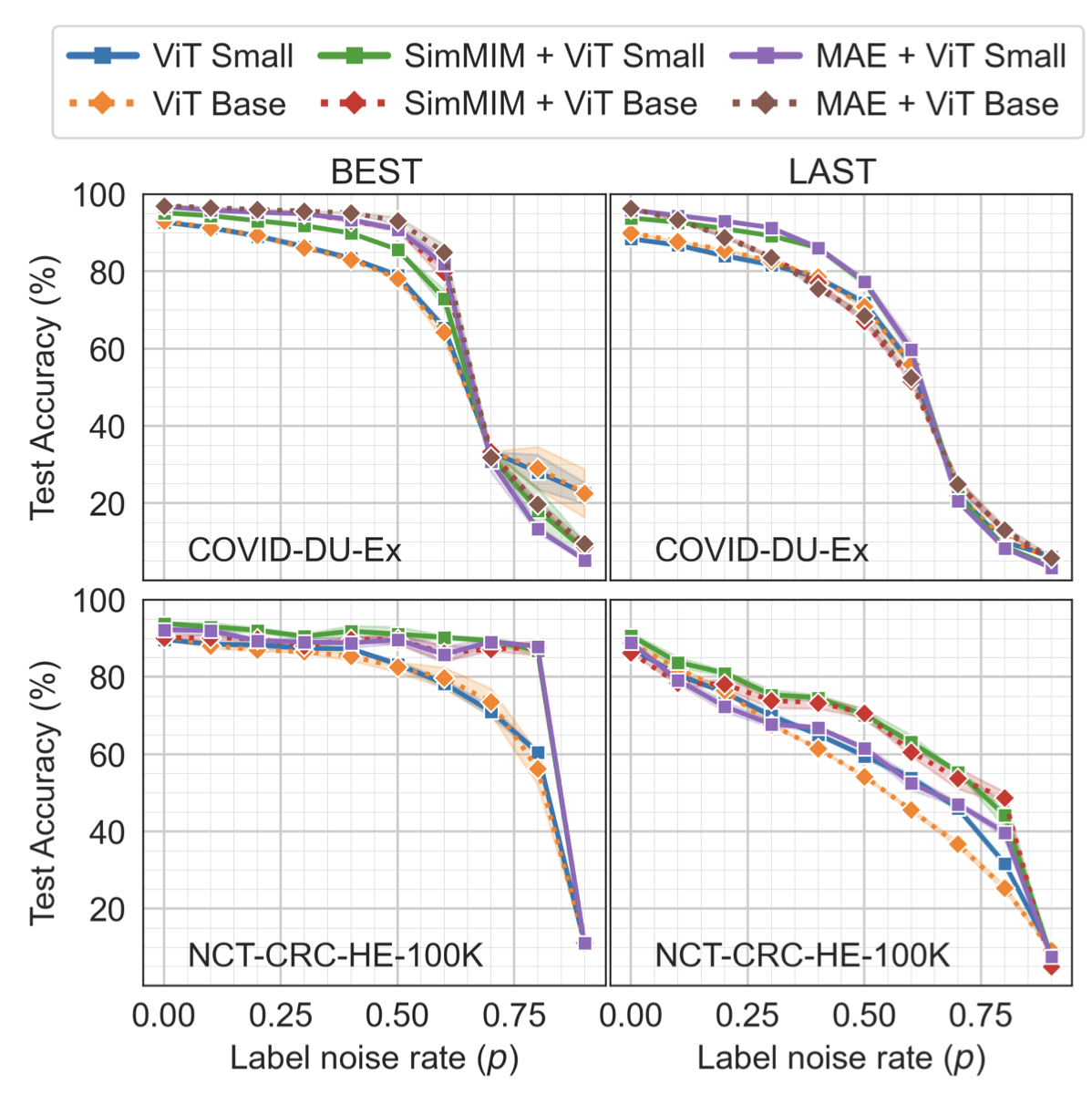
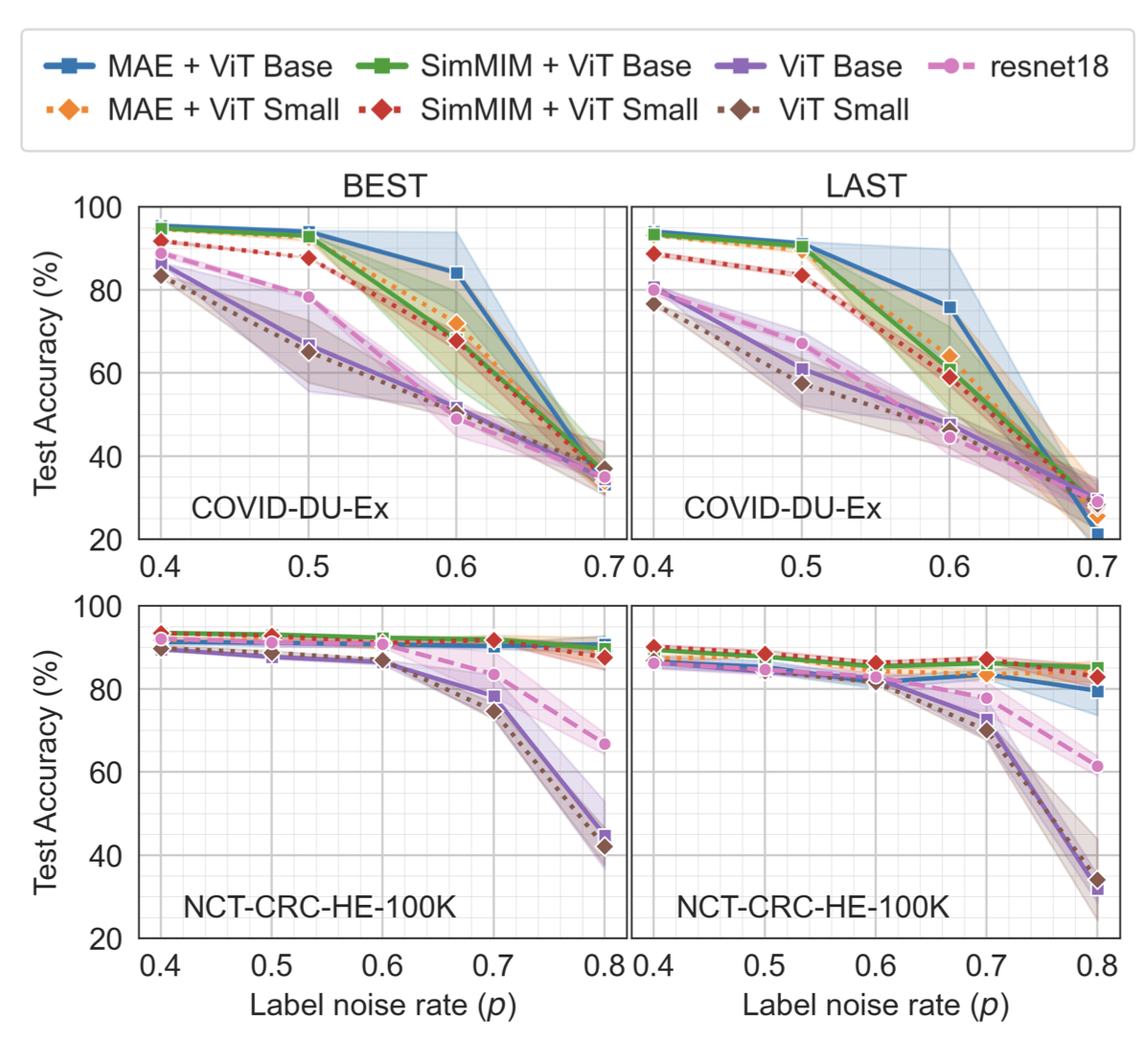
Label noise in medical image classification datasets significantly hampers the training of supervised deep learning methods, undermining their generalizability. The test performance of a model tends to decrease as the label noise rate increases. Over recent years, several methods have been proposed to mitigate the impact of label noise in medical image classification and enhance the robustness of the model. Predom- inantly, these works have employed CNN-based architectures as the backbone of their classifiers for feature extraction. However, in recent years, Vision Transformer (ViT)-based backbones have replaced CNNs, demonstrating improved performance and a greater ability to learn more generalizable features, especially when the dataset is large. Nevertheless, no prior work has rigorously investigated how transformer-based backbones handle the impact of label noise in medical image classification. In this paper, we investigate the architectural robustness of ViT against label noise and compare it to that of CNNs. We use two medical image classification datasets—COVID-DU-Ex, and NCT-CRC-HE-100K—both corrupted by injecting label noise at various rates. Additionally, we show that pretraining is crucial for ensuring ViT’s improved robustness against label noise in supervised training.
Improving Medical Image Classification in Noisy Labels Using only Self-supervised Pretraining
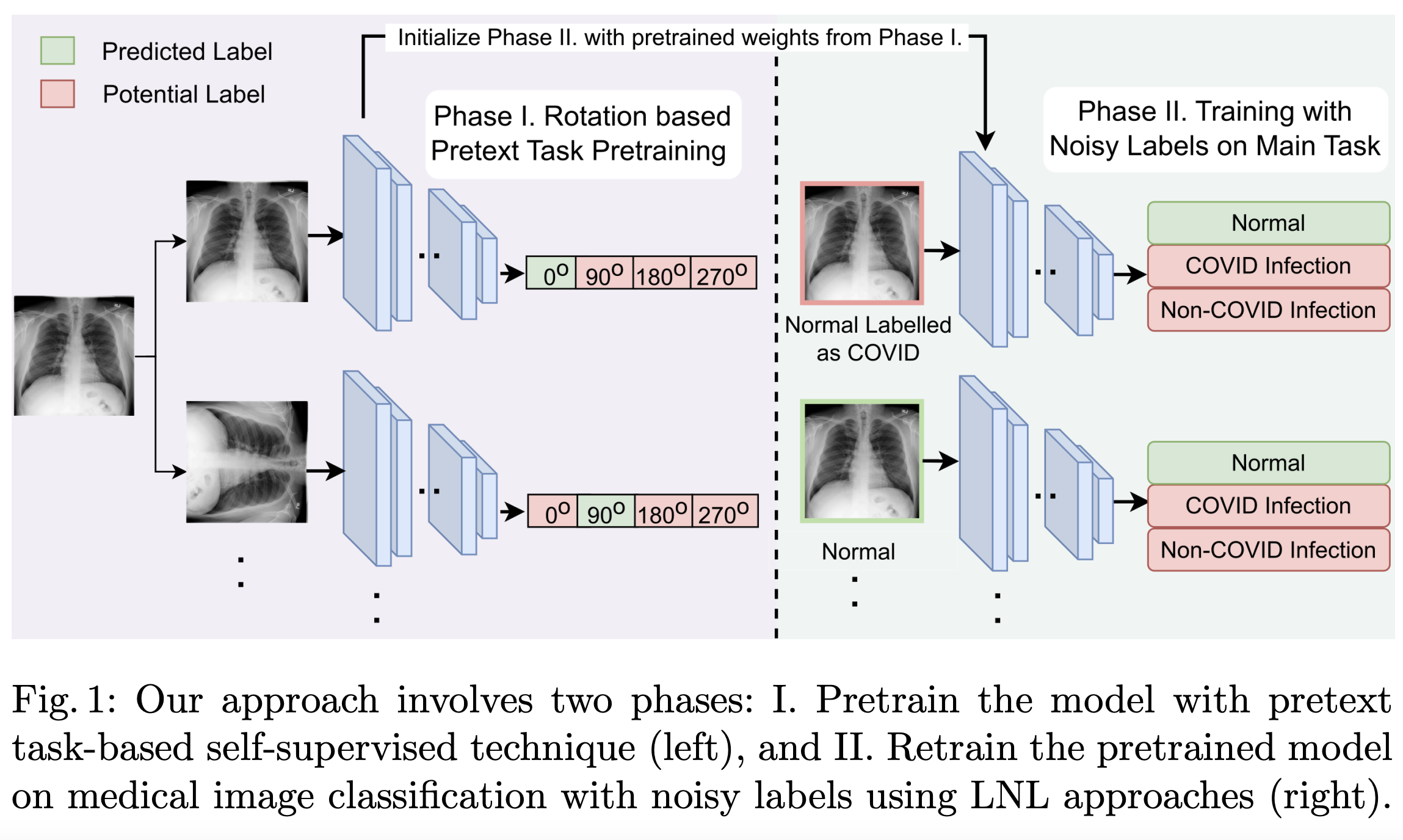
Noisy labels hurt deep learning-based supervised image classification performance as the models may overfit the noise and learn corrupted feature extractors. For natural image classification training with noisy labeled data, model initialization with contrastive self-supervised pretrained weights has shown to reduce feature corruption and improve classification performance. However, no works have explored: i) how other self-supervised approaches, such as pretext task-based pretraining, impact the learning with noisy label, and ii) any self-supervised pretraining methods alone for medical images in noisy label settings. Medical images often feature smaller datasets and subtle inter-class variations, requiring human expertise to ensure correct classification. Thus, it is not clear if the methods improving learning with noisy labels in natural image datasets such as CIFAR would also help with medical images. In this work, we explore contrastive and pretext task-based self-supervised pretraining to initialize the weights of a deep learning classification model for two medical datasets with self-induced noisy labels—NCT-CRC-HE-100K tissue histological images and COVID-QU-Ex chest X-ray images. Our results show that models initialized with pretrained weights obtained from self-supervised learning can effectively learn better features and improve robustness against noisy labels.
M-VAAL: Multimodal Variational Adversarial Active Learning for Downstream Medical Image Analysis Tasks
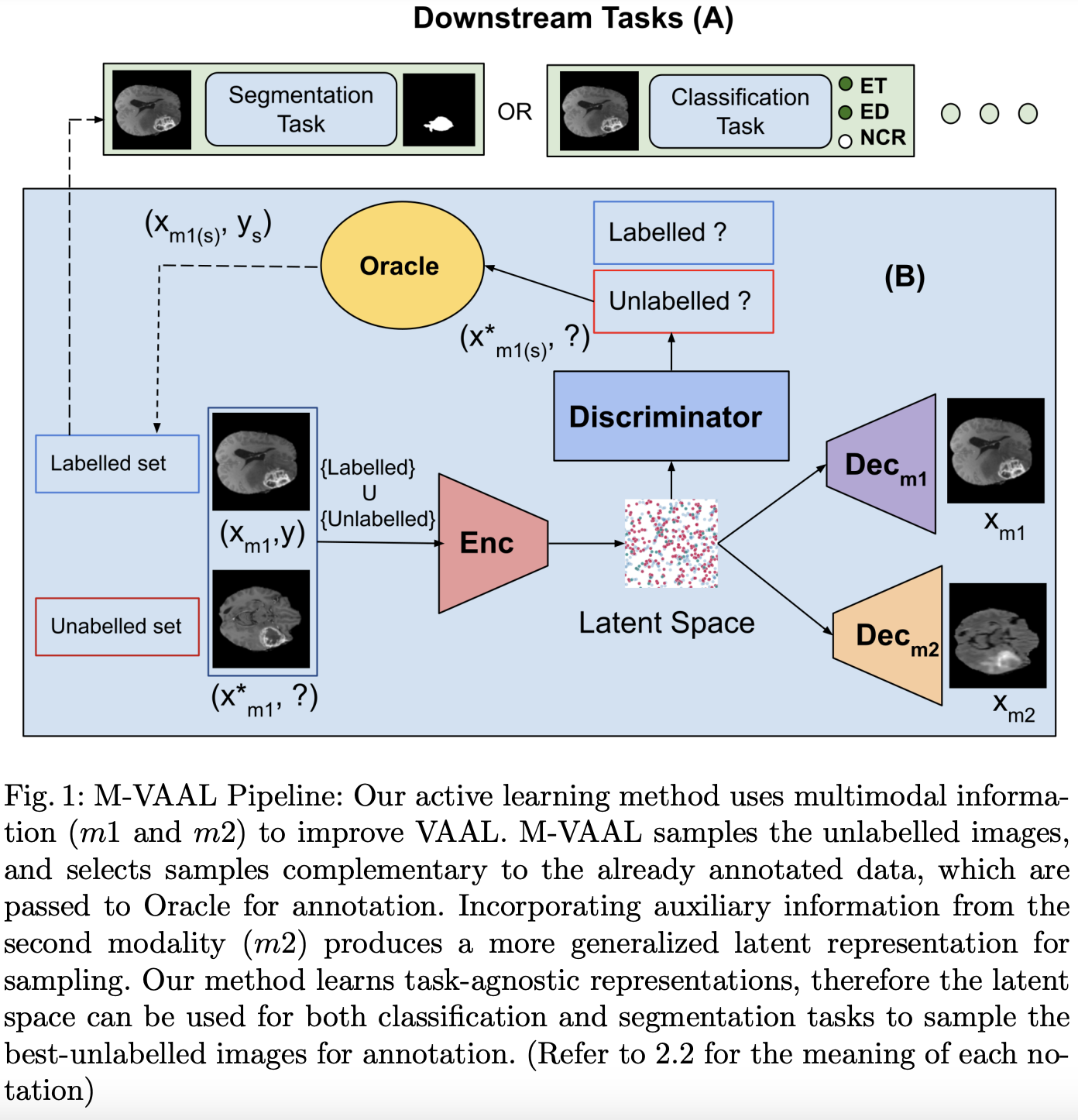
Acquiring properly annotated data is expensive in the medical field as it requires experts, time-consuming protocols, and rigorous validation. Active learning attempts to minimize the need for large annotated samples by actively sampling the most informative examples for annotation. These examples contribute significantly to improving the performance of supervised machine learning models, and thus, active learning can play an essential role in selecting the most appropriate information in deep learning-based diagnosis, clinical assessments, and treatment planning. Although some existing works have proposed methods for sampling the best examples for annotation in medical image analysis, they are not task-agnostic and do not use multimodal auxiliary information in the sampler, which has the potential to increase robustness. Therefore, in this work, we propose a Multimodal Variational Adversarial Active Learning (M-VAAL) method that uses auxiliary information from additional modalities to enhance the active sampling. We applied our method to two datasets: i) brain tumor segmentation and multi-label classification using the BraTS2018 dataset, and ii) chest X-ray image classification using the COVID-QU-Ex dataset. Our results show a promising direction toward data-efficient learning under limited annotations.
Investigating the impact of class-dependent label noise in medical image classification
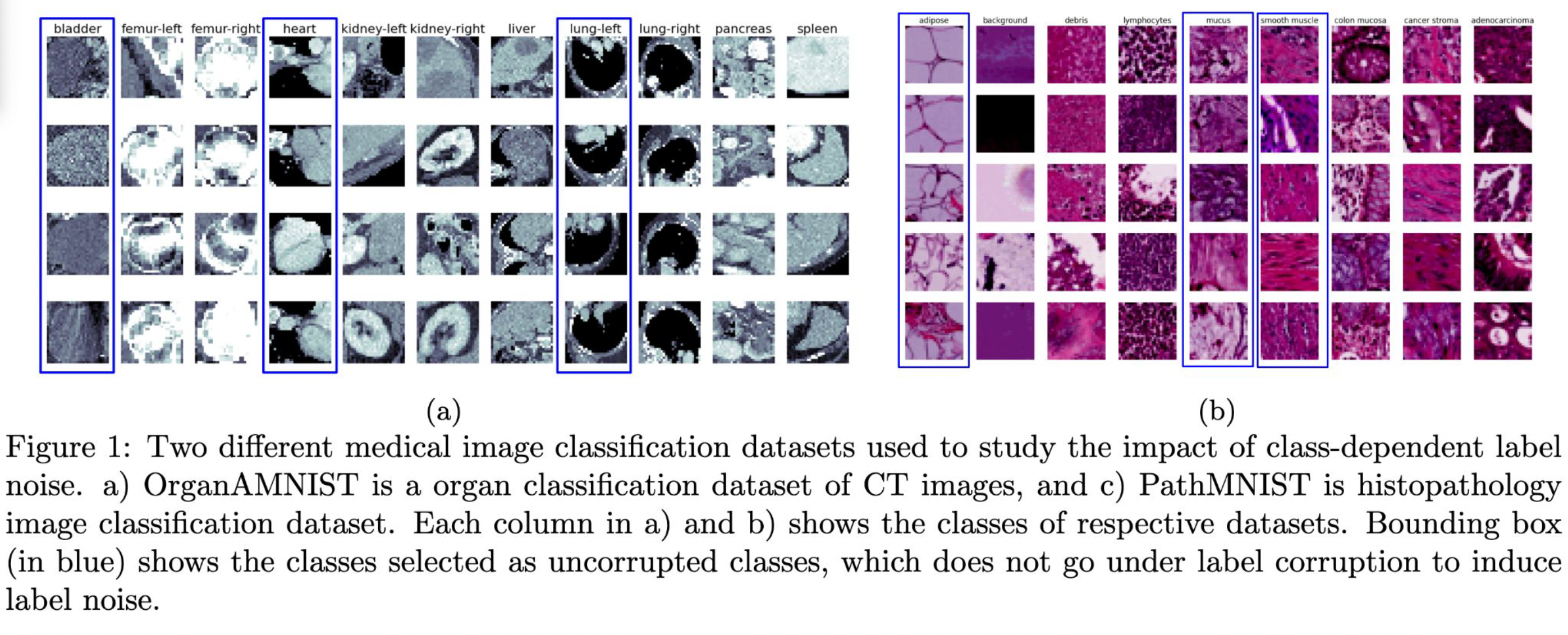
Label noise is inevitable in medical image databases developed for deep learning due to the inter-observer variability caused by the different level of expertise of the experts annotating the images, and, in some cases, the automated methods that generate labels from medical reports. It is known that incorrect annotations or label noise can degrade the actual performance of supervised deep learning models and can bias the models evaluation. Existing literature show that noise in one class has minimal impact on model’s performance for another class in natural image classification problems where different target classes have relatively distinct shape and share minimal visual cues for knowledge transfer among the classes. However, it is not clear how class-dependent label noise affects the model’s performance when operating on medical images, for which different output classes can be difficult to distinguish even for experts, and there is a high possibility of knowledge transfer across classes during the training period. We hypothesize that for medical image classification tasks where the different classes share very similar shape with differences only in texture, the noisy label for one class might affect the performance across other classes unlike the case when the target classes have different shape and are visually distinct. In this paper, we study this hypothesis using two publicly available datasets: a 2D organ classification dataset with target organ classes being visually distinct, and a histopathology image classification dataset where the target classes look very similar visually. Our results show that the label noise in one class has much higher impact on the model’s performance on other classes for histopathology dataset compared to the organ dataset.
How Does Heterogeneous Label Noise Impact Generalization in Neural Nets?
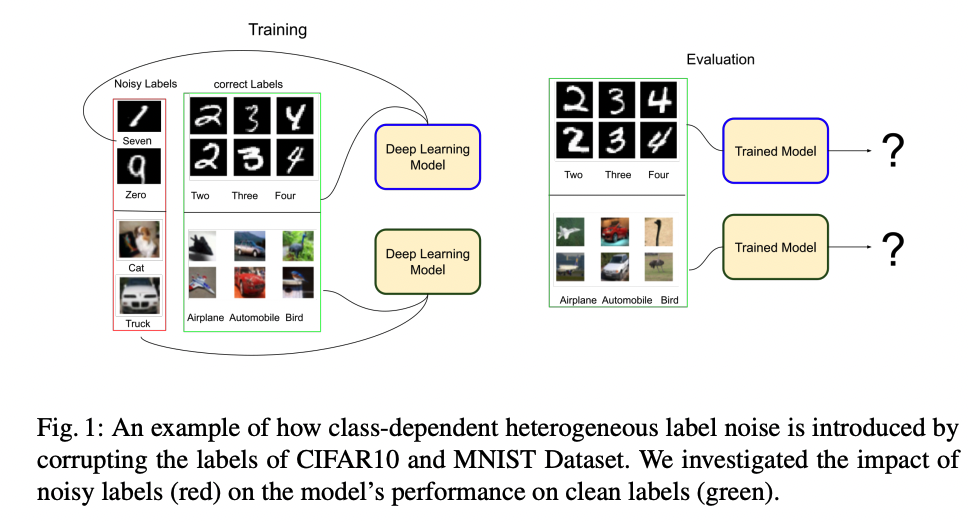
Incorrectly labeled examples, or label noise, is common in real-world computer vision datasets. While the impact of label noise on learning in deep neural networks has been studied in prior work, these studies have exclusively focused on homogeneous label noise, i.e., the degree of label noise is the same across all categories. However, in the real-world, label noise is often heterogeneous, with some categories being affected to a greater extent than others. Here, we address this gap in the literature. We hypothesized that heterogeneous label noise would only affect the classes that had label noise unless there was transfer from those classes to the classes without label noise. To test this hypothesis, we designed a series of computer vision studies using MNIST, CIFAR-10, CIFAR100, and MS-COCO where we imposed heterogeneous label noise during the training of multi-class, multi-task, and multi-label systems. Our results provide evidence in support of our hypothesis: label noise only affects the class affected by it unless there is transfer.
Efficient Online Continual Learning
Worked on developing efficient online learning classifier that is capable of learning from single pass through the dataset while being computationally efficient. For this work, we proposed a large-margin regularized multi-linear discriminant analysis algorithm which can dynamically create and collapse modes observed in the stream of data.
Label Geometry Aware Discriminator for Conditional Generative Networks

Multi-domain image-to-image translation with conditional Generative Adversarial Networks (GANs) can generate highly photo realistic images with desired target classes, yet these synthetic images have not always been helpful to improve downstream supervised tasks such as image classification. Improving downstream tasks with synthetic examples requires generating images with high fidelity to the unknown conditional distribution of the target class, which many labeled conditional GANs attempt to achieve by adding soft-max cross-entropy loss based auxiliary classifier in the discriminator. As recent studies suggest that the soft-max loss in Euclidean space of deep feature does not leverage their intrinsic angular distribution, we propose to replace this loss in auxiliary classifier with an additive angular margin (AAM) loss that takes benefit of the intrinsic angular distribution, and promotes intra-class compactness and inter-class separation to help generator synthesize high fidelity images. We validate our method on RaFD and CIFAR-100, two challenging face expression and natural image classification data set. Our method outperforms state-of-the-art methods in several different evaluation criteria including recently proposed GAN-train and GAN-test metrics designed to assess the impact of synthetic data on downstream classification task, assessing the usefulness in data augmentation for supervised tasks with prediction accuracy score and average confidence score, and the well known FID metric.
Spine Curvature Estimation from X-ray Images
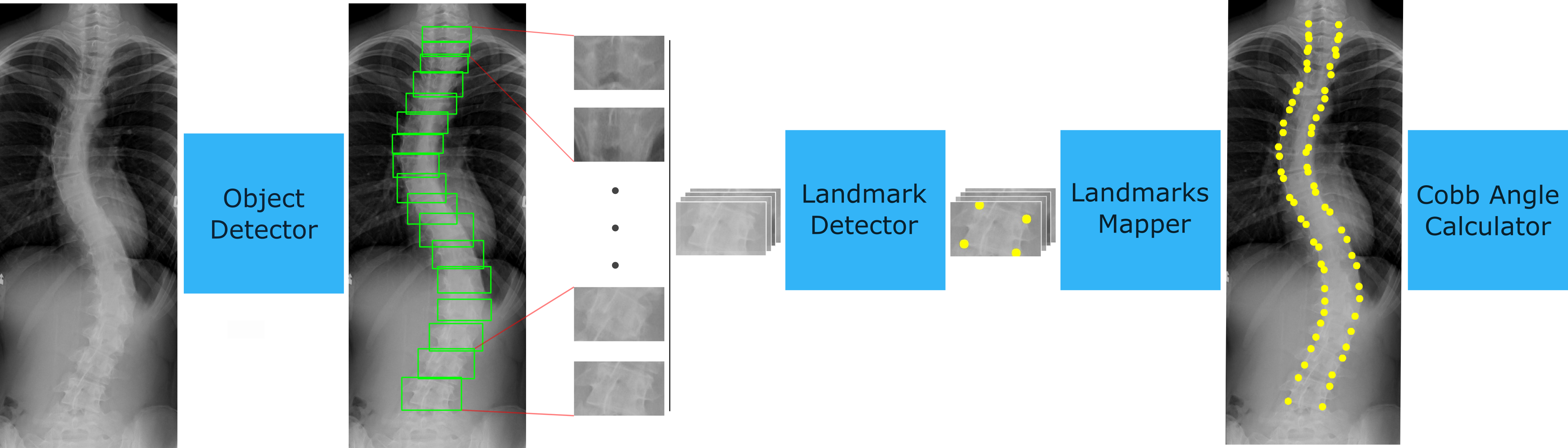
Correct evaluation and treatment of Scoliosis require accu- rate estimation of spinal curvature. Current gold standard is to manually estimate Cobb Angles in spinal X-ray images which is time consuming and has high inter-rater variability. We propose an automatic method with a novel framework that first detects vertebrae as objects followed by a landmark detector that estimates the 4 landmark corners of each vertebra separately. Cobb Angles are calculated using the slope of each vertebra obtained from the predicted landmarks. For inference on test data, we perform pre and post processings that include cropping, outlier rejection and smoothing of the predicted landmarks. The results were as- sessed in AASCE MICCAI challenge 2019 which showed a promise with a SMAPE score of 25.69 on the challenge test set.
With this work, we participated in Accurate Automated Spinal Curvature Estimation MICCAI 2019 Challenge.
Estimating Pesticide Concentration with Smartphone
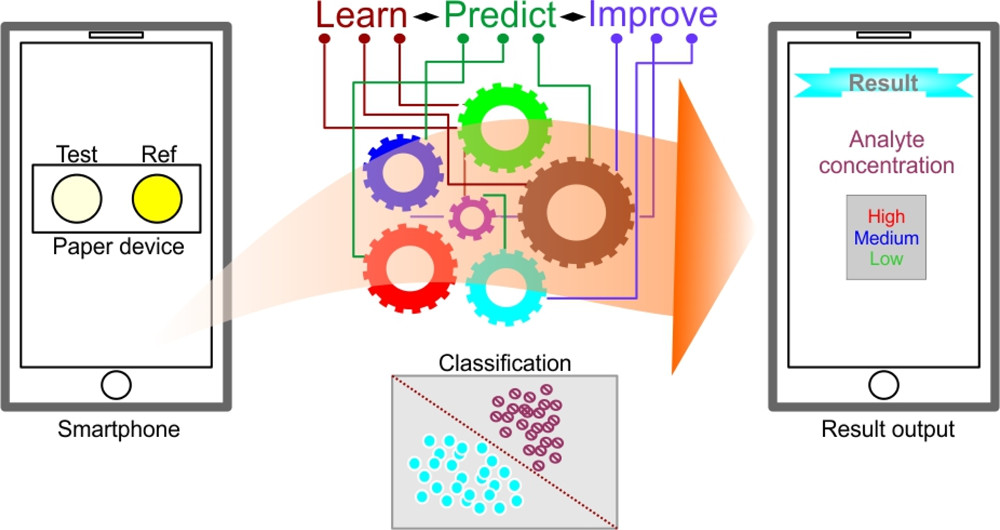
Paper-based analytical devices (PADs) employing colorimetric detection and smartphone images have gained wider acceptance in a variety of measurement applications. PADs are primarily meant to be used in field settings where assay and imaging conditions greatly vary, resulting in less accurate results. Recently, machine-learning (ML)-assisted models have been used in image analysis. We evaluated a combination of four ML models-logistic regression, support vector machine (SVM), random forest, and artificial neural network (ANN)-as well as three image color spaces, RGB, HSV, and LAB, for their ability to accurately predict analyte concentrations. We used images of PADs taken at varying lighting conditions, with different cameras and users for food color and enzyme inhibition assays to create training and test datasets. The prediction accuracy was higher for food color than enzyme inhibition assays in most of the ML models and color space combinations. All models better predicted coarse-level classifications than fine-grained concentration classes. ML models using the sample color along with a reference color increased the models’ ability to predict the result in which the reference color may have partially factored out the variation in ambient assay and imaging conditions. The best concentration class prediction accuracy obtained for food color was 0.966 when using the ANN model and LAB color space. The accuracy for enzyme inhibition assay was 0.908 when using the SVM model and LAB color space. Appropriate models and color space combinations can be useful to analyze large numbers of samples on PADs as a powerful low-cost quick field-testing tool.
Synthetic to Real Domain Translation Using Conditional GAN
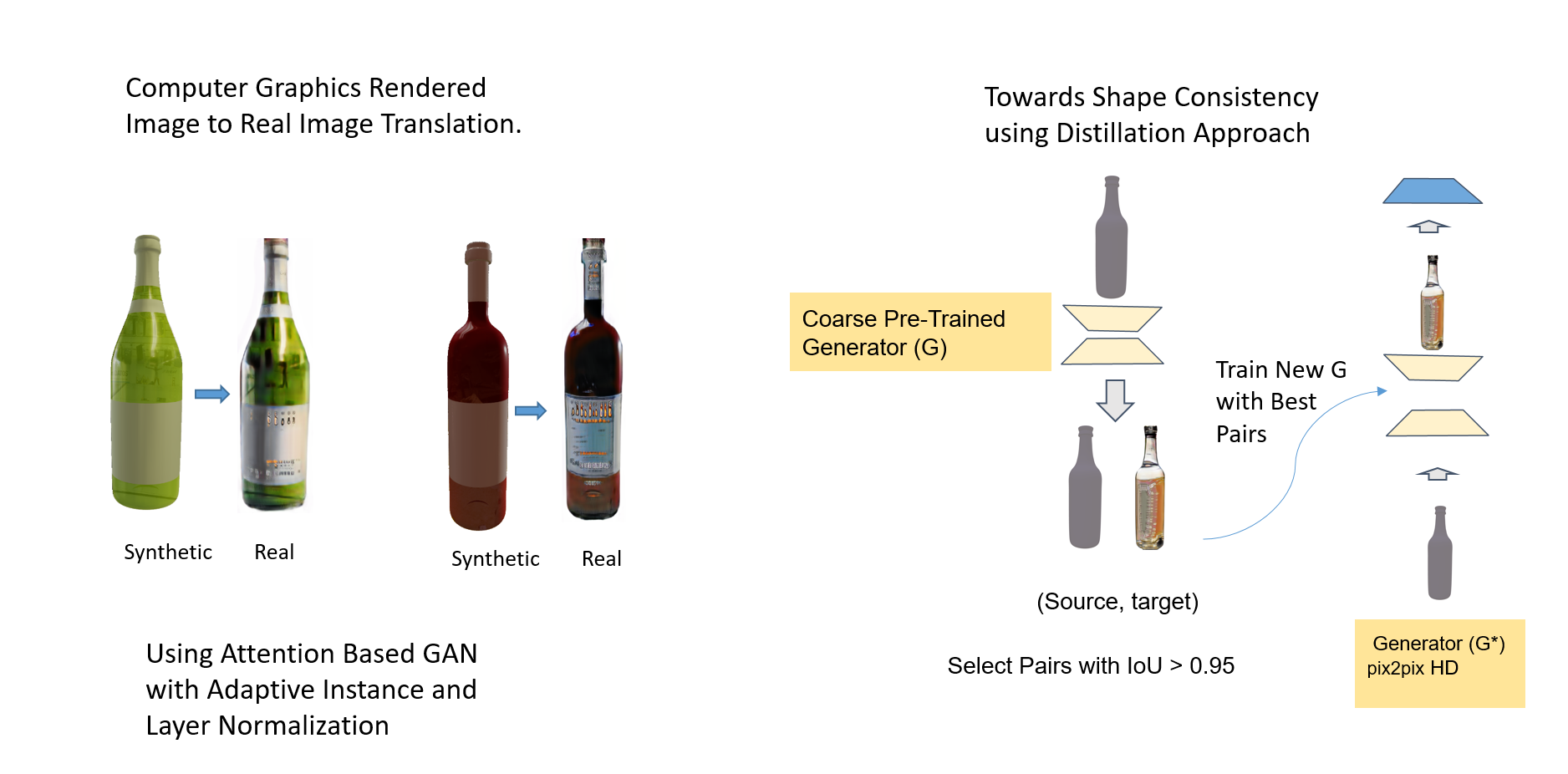
In this project, we looked into the problem with the conditional GANs in preserving the shape of the imput image. We solved this problem by using two GANs. First GAN trains with unpaired images. The best generated images are distilled using IOU criterion, thus generating good pairs of images. These pairs are then use to train a second GAN network (pix2pixHD). The later network trained with distilled examples learn to preserve the shape of the input.
